Mitomycin C (MC) (structure 1) is a potent antitumor drug. MC, in combination with other antitumor drugs, is often used clinically for the treatment of advanced breast cancer. Two closely related derivatives of MC, namely, 10-decarbamoyl mitomycin C (2; DMC) and 2,7-diaminomitosene (3; 2,7-DAM) and their DNA adducts are also relevant to the effects of MC.
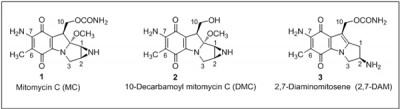
Cellular toxicity of 2,7-DAM is marginal and it does not activate the p53 pathway, whereas MC and DMC are highly cytotoxic and are known to activate the p53 pathway. DMC exhibits higher toxicity than MC and is lethal to p53-deficient cells by inducing degradation of Checkpoint 1 protein, which was not observed following MC treatment of the p53-deficient cells. This dissimilarity in the cell death pathways activated by the MC and DMC is thought to be due to differential signaling by the DNA adducts.
MC forms several adducts with DNA. In cells MC undergoes reductive activation followed by covalent DNA binding with deoxyguanosine residues to form a monoadduct. Subsequently, it can form an intra-strand cross-link at GG- sites or an inter-strand cross link at a CG- site. These DNA adducts, if are not repaired, can block transcription and DNA replication and cause cell death. The antitumor activity of MC is generally believed to be derived from its interactions with DNA that eventually cause cell death.
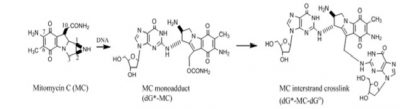
Both the monadducts and the cross-links of MC are substrates for repair by the nucleotide excision repair proteins, such as UvrABC of E. coli. We determined that UvrABC proteins generate three different types of incisions in DNA near the MC cross-link sites. Each of these double strand breaks would be lethal.
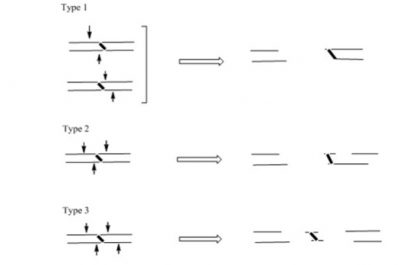
Evidently, addressing additional questions on the biological effects of DNA-DNA inter-strand cross-links like this one and by other antitumor agents are critical to design and synthesize new drugs.